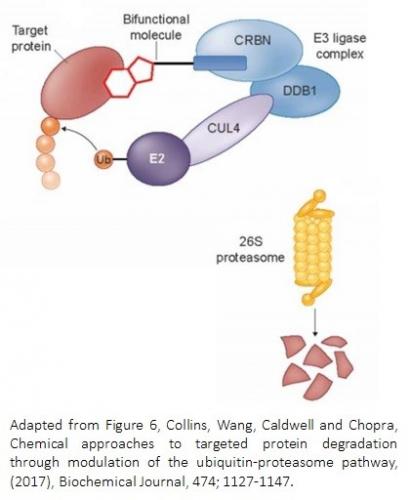
YCMD is introducing a proprietary, new small molecule library of chemical degraders for cell-based screening. Like the PROTAC approach, these library molecules have two binding regions. The constant binding portion binds to a component of cellular degradation machinery (i.e. cereblon, substrate recognition component of an E3 protein ligase complex). The variable binding portion alters the surface of cereblon to enable interaction with countless new protein substrates. This library is ideal for screening the complex biology of cellular systems. Small changes in chemical structure alter substrate specificity, and when a new protein substrate binds with sufficient affinity, it will be degraded, resulting in a phenotypic change for essential functions.
Following the screen, a subset of hits will be confirmed for degradation-dependence by synthesizing and testing cereblon-resistant analogs.
Advantages of this chemical degrader library include:
- Novel ligands for “undruggable” proteins
- Low molecular mass compounds facilitate cellular permeability
- Degradation of family members from a conserved ligand-binding domain increases the likelihood of observing a phenotype
We have set aside pilot funds to alpha test this library free-of-charge for screening assays involving cell lines that meet the following criteria:
- Assays for paired human cell lines differing in the expression of a single target, such as a knock-out, over-expressed or mutated target for use in proliferation assays. With this phenotypic approach, differential activity observed between cell lines can map compound activity to a pathway.
- Assays using paired cell lines quantifiable by a platereader using other phenotypic endpoints.
- Optimized high content image-based assay scaled to 96- or 384-well format. Analysis algorithm for feature extraction (segmentation) and quantitation in InCell Analyzer (GE) or CellProfiler format. We will entertain requests for optimized high content (image-based) assays on a limited basis.
Please answer the following questions for your application:
- Describe the pathway and its connection to biological processes.
- What is the target (or target pathway) and describe the cell lines used for screening (species, tissue of origin, differences in construct between paired lines, promoters)?
- What are the culture conditions for your cell line(s) (media, supplements, inducers)?
- Describe the assay method and readout and efforts to optimize the assay for plate-based format to date (if any).
Send questions and completed applications to yulia.surovtseva@yale.edu. Applications will be reviewed on a rolling basis.